Mechanical clamps in proteins are defined as a structural region which is responsible for the biggest contribution to mechanostability. We determine the mechanical clamps by identifying contacts that break near the largest force peak and then studying dynamical effects associated with a removal of various groups of these contacts. This task is accomplished only for selected proteins.
We divide the mechanical clamps into two main groups: I. involving small strains in localized regions (usually in secondary structures) and II. involving a larger motion of at least one loop made of a segment of the backbone.
Group I: Strain in localized regions
Examples of mechanical clamps belonging to the first group are shown in Figure 1 where the black arrow usually stands for a ¯-strand but sometimes it may have a more generic meaning – it can also indicate a helix. We distinguish the following motifs:
Ia. Elementary motifs- S – shearing between parallel β-strands (it may also take place between two helices.
- SA – shearing between atiparallel β-strands (including the hairpins).
- Z – unzipping of two β-strands.
- Za – unzipping of two helices.
- U – unstructured: shearing between nearby unstructured segments of the backbone.
Ib. Composite motifs
They consist of several elementary motifs
- SD1 – disconnected: S is followed by S, separated by an essentially strain free region.
- SD2 – disconnected: SA is followed by SA.
- SD3 – disconnected: S is followed by SA.
- SS – supported: the main S motif is flanked by neighboring β-strands which stabilize it.
Shearing the main S results also in a shear with the flanking β-strands.
- T – torsion: multiple S motifs arranged in a bent structure (if the resistance to stretching
comes from the links between the S motifs).
- SB – shear box: a sheared two-strand β-sheet that is placed below a sheared two-helix
'sheet' so that a shear in one strand induces a shear on the helix above.
- D – a delocalized clamp with multiple elementary clamps exerting comparable resistances to pulling. Some regions can by unstructured.
Group II: Mechanical clamps involving backbone loops
Examples of mechanical clamps belonging to the second group are shown in Figure 2. In most of thesea, the loops arise due to the presence of disulphide bonds, indicated by short solid lines.
- CK – the cystine knot: shearing takes place inside a cystine knot – a loop that is
created by two disulphide bonds.
- CL1 – the cystine loop: shearing results because one branch of S is pulled by a cystine
loop.
- CL2 – the cystine loop: similar to CL1 but the motion of the cystine loop also drags
another piece of the backbone that transmits shear to the other branch of the S motif.
- CSK – the cystine slipknot motif: involves three disulphide bridges which generate two
loops: knot-loop (which is an example of a cystine knot) and a slip-loop. The mechanical
resistance to pulling comes from pulling the slip-loop through the knot-loop.
- SK – the slipknot motif: created by two interacting loops, the slip-loop and the knotloop, that move simulateneously on pulling. If the knot loops shrinks faster than the knot loop then the slipknot gets tightened temporarily and a ”catch bond” is formed. This intermediate and metastable configuration eventually gets untied upon further stretching.
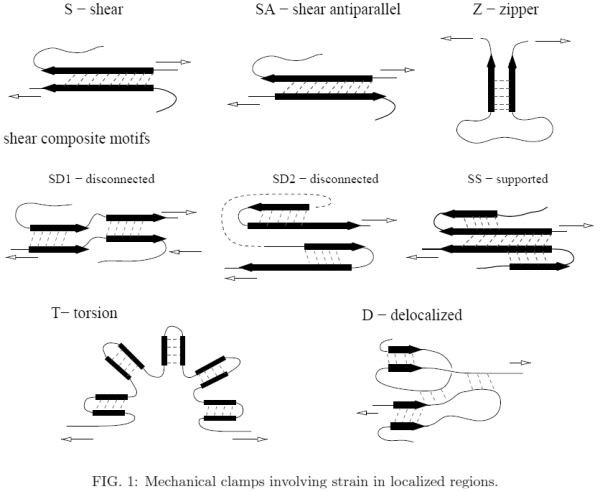
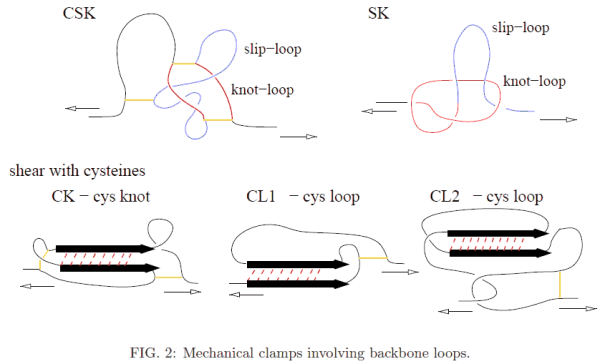
TABLE I: Examples of mechanical clamps - discussed in the literature. The references are provided in the tables summarizing experimental and all-atom data in the BSDB.
| PDB | motifs | protein | PDB | motifs | protein |
| 1g1c | SD2 | I1(reduced) | 1tit | S | I27 |
| 1oww | SD2 | 1FNIII | 1fnf | SD2 | 7FNIII |
| 1fnh | SD2 | 10FNIII | 1fnh | SD2 | 12FNIII |
| 1ten | SD2 | 13FNIII TNFNAll | 1nct | S | I54-I59 |
| 1aj3 | Z | α-spectrin | 1s35 | Z | β-spectrin1−4 |
| 1cfc | Z | calmodulin | 1qjo | Z | E2lip3 |
| 1n11 | Z | ankyrin*1 | 1n11 | Z | ankyrin*24 |
| 1edh | S | cad1 | 1edh | S | cad2 |
| 1vsc | S | cell adhesion VCAM1 | 1vsc | SD2 | cell adhesion VCAM2 |
| 1ksr | Z | FLN4 | 1dqv | Z | C2A |
| 1b6i | Za | T4 lysozyme | 1bnr | Z | barnase |
| 1j85 | S | methylotransferase | 1ubq | SS | ubiquitin(48-C) |
| 1ubq | SS | ubiquitin(N-C) | 1ubq | Z | ubiquitin(11-C) |
| 1emb | SD1 | GFP(3-212) | 1emb | Z | GFP(132-212) |
| 1emb | ss | GFP(3-132) | 1emb | SD1 | GFP(117-182) |
| 1pga | SS | GB1 | 1hz6 | SS | protein L |
| 1anu | SD1 | scaffoldin c2A | 1aoh | SD1 | scaffoldin c7A |
| 1g1k | SD1 | scaffoldin c1C | 1v9e | CK | BCA II |
| 2o9c | S | phytochrome | 2o9c | S | phytochrome* |
TABLE II: Examples of proteins with small experimental values of Fmax (< 40 pN) as discussed in the literature.
| PDB | motifs | protein |
| 1cyi | no Fmax | cc6 |
| 1bdd | no Fmax | 1gb |
| 1rsy | no Fmax | c2 |
TABLE III: Examples of mechanical clamps, as assessed based on our model
| PDB | motifs | PDB | motifs | PDB | motifs | PDB | motifs |
| 1bmp | CSK | 1qty | CSK | 2bhk | CSK | 1lxi | CSK |
| 1cz8 | CSK | 2gh0 | CSK | 1wq9 | CSK | 1flt | CSK |
| 1fzv | CSK | 2gyz | CSK | 1rew | CSK | 1m4u | CSK |
| 1vpf | CSK | 1c4p | SS | 1qqr | SS | 3bmp | CSK |
| 1j8s | SS | 1wq8 | CSK | 1j8r | SS | 1f3y | SS |
| 2vpf | CSK | 2h64 | CSK | 1kdm | CL2 | 1q56 | CL2 |
| 1rv6 | CSK | 1waq | CSK | 1reu | CSK | 1tgj | CSK |
| 2pbt | SS | 2h62 | CSK | 1tgk | CSK | 2fzl | SS |
| 1qu0 | CL2 | 1f5f | CL2 | 1dzk | CL2 | 1aoh | SS |
| 1vsc | CL2 | 2c7w | CSK | 2gyr | CSK | 1dzj | CL2 |
| 2sak | SS | 2bzm | CL2 | 2g7i | CL2 | 1i04 | CL2 |
| 1hqp | CL2 | 1afk | CL1 | 1qoz | CL1 | 1lx5 | CYS |
| 1oxm | CL2 | 1hfh | CL1 | 1afl | CL1 | 1aqp | CL1 |
| 2g4x | CL1 | 2g4w | CL1 | 2a2g | CL2 | 2boc | CL2 |
| 1vpp | CSK | 1lm8 | SA | 1ppx | SD2 | 1jrk | SD2 |
| 1k26 | SD2 | 1pun | SD2 | 1pus | SD2 | 1oo2 | SD2 |
| 1ssn | SD2 | 1i3v | SD2 | 1pav | SB | 1qp1 | D |
| 1amx | SA | 1ei5 | SA | 1vhp | D | 1tum | D |
| 1f86 | D | 1ui9 | D | 1pqe | D | 1mg4 | D |
| 1lsl | D | ||||||

